Research at the Institute for Biological Physics, in CRC 1310, and in the Center for Data and Simulation Science.
Antibiotic resistance and drug combinations
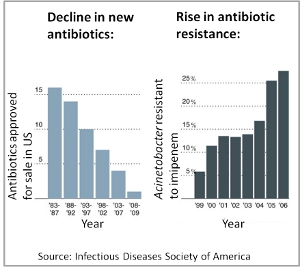
The rapid rise of antibiotic resistance is a serious concern for public health, in particular since the discovery of new antibiotics has essentially come to a halt. How can this looming crisis be averted? We aim to find new ways of combining drugs and other compounds to minimize or even prevent the evolution of drug resistance. As a prerequisite we aim to elucidate the underlying causes of drug interactions such as synergism and antagonism.
Further reading:
- Nature Reviews Microbiology 2022
- Current Opinion in Systems Biology 2021
- PLOS Computational Biology 2021
- Nature Communications 2020, press release
- Nature Communications 2020, press release, HFSP news
- Die Presse (in German)
- Current Opinion in Biotechnology 2017
- PLOS Biology 2015
- Molecular Systems Biology 2015
- Current Opinion in Microbiology 2015
- Environmental Microbiology Reports 2014
- Chemistry & Biology 2014
- Cell 2009; Perspective in New England Journal of Medicine by Xavier and Sander
- ACS Chemical Biology 2009
- Our perspective on Davies et al., Molecular Cell 2009
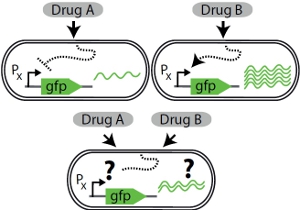
Cellular responses to drug signals
Cells are often faced with the simultaneous or sequential presence of multiple signals that individually elicit opposite gene expression responses. How do cells resolve such gene regulatory conflicts? We use fluorescent reporters, time-lapse microscopy combined with microfluidics, and a state-of-the-art robotic system to reveal how the bacterium Escherichia coli and the budding yeast Saccharomyces cerevisiae respond to combinations of antimicrobial drugs and other stressors. Antimicrobials are important drugs for the treatment of infectious diseases. They are often used in combination, making them a particularly relevant example.
Further reading:
Microbial communities and polymicrobial infections
Many infections involve more than one bacterial species. How do bacteria in such polymicrobial infections interact with each other? Do these interactions affect the sensitivity of the bacterial community to antibiotics? We systematically map quantitative ecological interaction networks for bacteria in polymicrobial infections and analyze them using the framework of statistical network theory. We further develop theoretical descriptions of these small ecosystems to understand and predict their dynamics.
Further reading:
- PLOS Computational Biology 2021
- PNAS 2017; press release (in German)
Theoretical models of bacterial growth
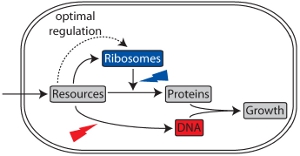
Even though the main cellular targets of many drugs are known, their effects on cell physiology and growth are often hard to rationalize. Are there quantitative laws governing bacterial growth in the presence of antibiotics? We develop a theoretical framework to describe bacterial growth and gene regulation. Of particular interest in these models is the regulation of cellular stress response and drug resistance systems. We use the toolbox of dynamical systems theory and statistical physics to analyze these models and perform experiments to test our predictions directly.
Further reading:
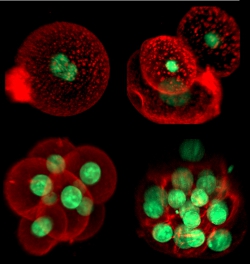
Theoretical models of cell differentiation, migration, and signaling
We are interested in cell differentiation, migration, communication between cells by secreted signaling molecules, and other key processes in the development and immune system of higher organisms such as fruit flies and mice. How do signaling molecules move through cells and tissues? How do cells accurately detect and respond to such signals? We build physics-inspired theoretical descriptions of cellular processes and compare them to experimental data using quantitative image analysis. We collaborate with developmental and cell biology labs to quantify key parameters and thus improve our understanding of these fascinating phenomena.
Further reading: